Projects
1 Why I stopped using Precision and Accuracy for binary classification
Precision and Accuracy are the most common performance metrics when dealing with binary classification problems. Doing some research, I found out that they are not robust enough for frequent scenarios in biomedical data science. Find out why, and what is the alternative metric on my LinkedIn article. All the code is available at this GitHub repository.

2 Shiny app to explore investments diversification
Diversification is essential in an investment portfolio. That is why I developed a Shiny app to explore the diversification of my investments in Microwd. All the code is available at this GitHub repository.
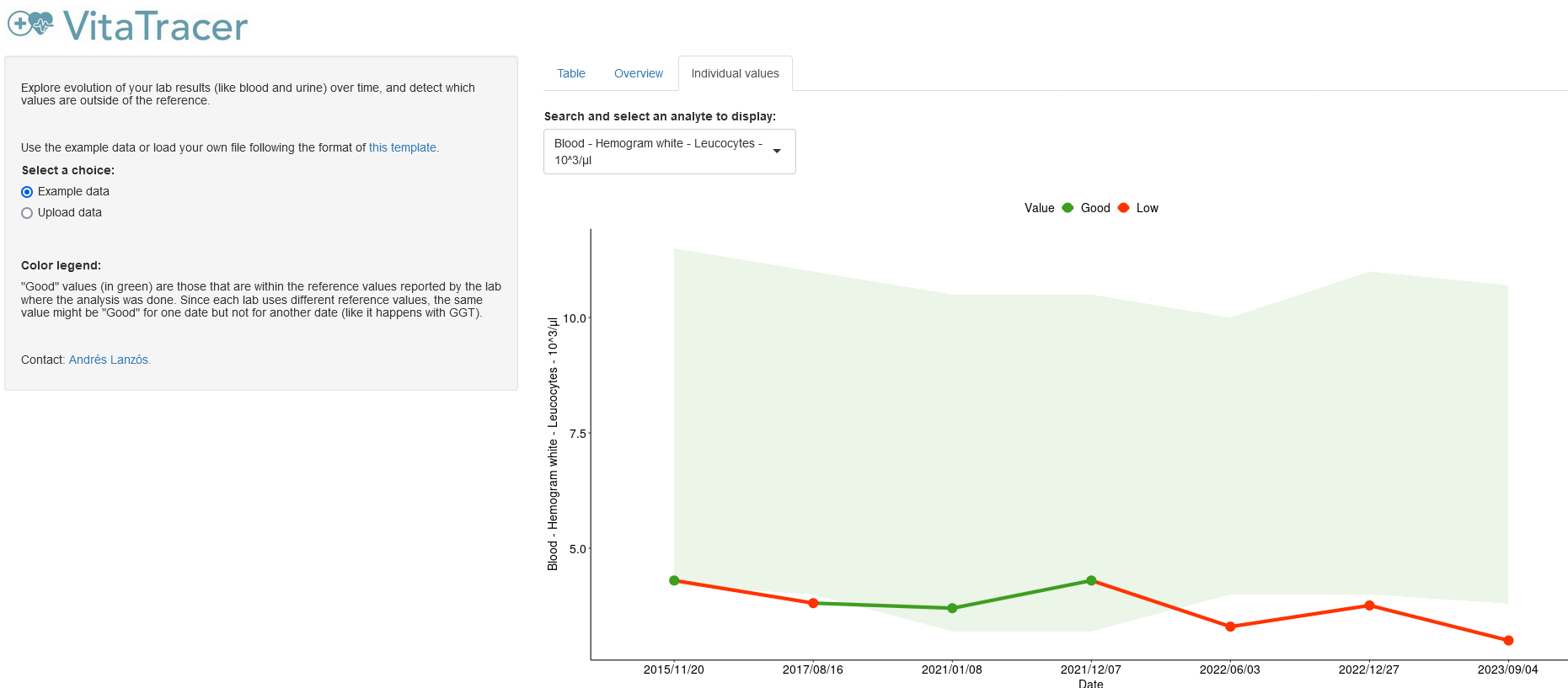
3 Shiny app to trace your blood/urine test
Every time I have done a blood or urine test, it was difficult to compare to previous values. That is why I collected my results from the last +5 years and created VitaTracer. This Shiny app allows me to see a summary of all my values, and inspect how they evolved over time. All the code is available at this GitHub repository.
4 Shiny app to explore the 3D structure of proteins measured by Mass Spectrometry
Back in 2021, I was working at R&D department of Biognosys when AlphaFold2 structures were released, and I realized that such a resource could be very useful for our company. I made a post with my ideas in our internal network, an a few days later the CEO of the company came to my desk to discuss it. After a couple months of work in top of my main project, we released the 3D Protein Explorer.
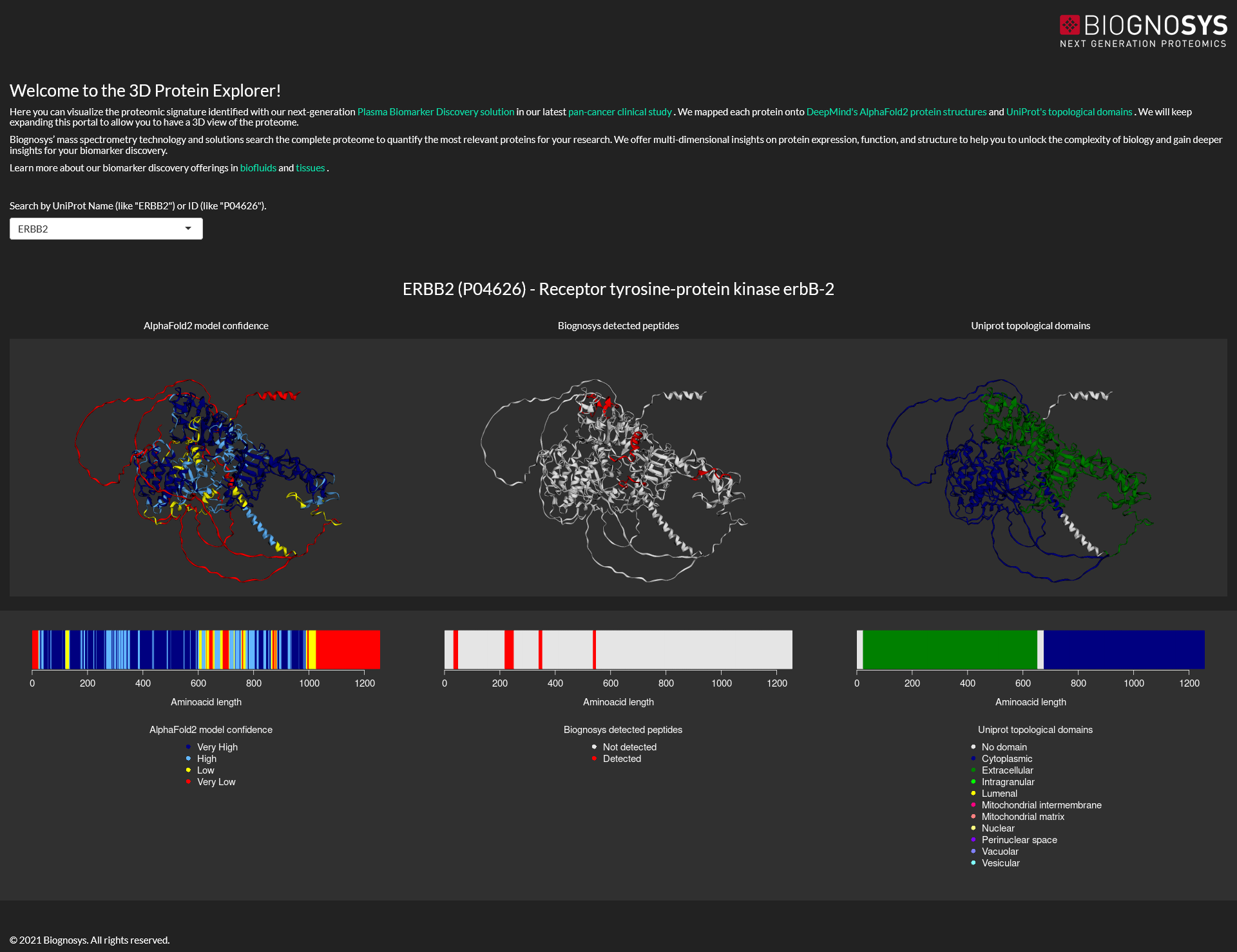
This was a great project to learn how to display the 3D structure of proteins and to create a Shiny app to be released to the public, two things that I never did before. While it’s a “simple” app, it was great to show to our customers which proteins we were able to detect in plasma samples, and exactly which regions we could see via Mass Spectrometry. The app is currently deployed on shinyapps.io, and I had to get creative to quickly display the protein that the user selected among thousands of them without freezing the app. It was a fun project where I collaborated with the business and marketing departments.
5 ExInAtor 1 & 2 - Finding cancer driver lncRNAs
I spent my PhD at the GOLD lab, under the supervision of Rory Johnson, in the search of cancer driver long noncoding RNAs (lncRNAs), a type of genes that are transcribed but do not encode proteins. During this time, I developed ExInAtor, the first tool ever designed to specifically find if lncRNAs were frequently mutated in cancer patients, hinting at a potential role in the onset of the disease (hence the name “cancer driver”). We published our findings at Scientific Reports, where I was first author.


As we learned more about these genes, I develop a second version, ExInAtor2, which included the functional impact of mutations, in addition to their frequency, to prioritize cancer driver lncRNAs. Additionally, Roberta Esposito led the experimental validation of ExInAtor2 predictions, and we published our findings in Nature Communications, where I was first co-author.
Both tools are a mix of Python and R code, and I had to work hard for ExInAtor2 to be able to run millions of simulations in a decent run time. Back in the days, there was no ChatGPT, so it was a fun experience to learn how to squish Python capabilities as a non-software-engineer.